
Artificial intelligence in healthcare—the road to precision medicine
Introduction
The existing paradigm of healthcare is based on the average-patient one-size-fits-all approach to deliver diagnostic, therapeutic and preventive interventions. However, it has become alarmingly clear that, while practical to implement, these broad clinical approaches fail to adequately address the medical needs of a significant portion of the population (1). When it comes to pharmacotherapy, for example, only 40–60% of patients respond to treatment (2,3). The variable drug treatment response rates now widely documented across medical literature are due to considerable inter-individual variability. A more targeted approach to medicine is thus clearly needed to optimise treatment, which has prompted the emergence of precision medicine. The terms precision medicine and personalised medicine have been used interchangeably, but the preferred current terminology is precision medicine, as there is a consensus that the term personalised medicine could be misinterpreted to imply that treatments and preventions are being developed uniquely for each individual. From an epidemiological, pharmacological and biological perspective, the scale of inter-individual and intra-individual variation in drug response as indicated above, it is pragmatic to assume that personalised treatment at an individual level is rather aspirational. Identifying and predicting subgroups with a better or worse response is likely more achievable and has a more realistic potential to revolutionise medicine.
In contrast to the one-size-fits-all model, precision medicine aims to integrate an individual’s unique features from clinical phenotypes and biological information obtained from imaging to laboratory tests and health records, to arrive at a tailored diagnostic or therapeutic solution with a higher chance of success (1). It is expected that patients will benefit from early accurate diagnosis, higher treatment efficacy and fewer adverse drug reactions, while broader improvements include greater healthcare savings and economic productivity. Precision medicine thus encompasses both diagnosis and prediction with greater accuracy than current clinical and epidemiological guidelines. The notion of precision medicine emerged from the dramatic successes in the identification of distinct subpopulations within certain cancer categories through advances in genomic sequencing followed by effective targeting of these molecular cancer subtypes by specific drugs. Patients with chronic myeloid leukaemia whose tumours harbour the BCR/ABL translocation (‘Philadelphia chromosome’) are successfully treated with the drug imatinib that inhibits tyrosine kinase (4); patients with cystic fibrosis can benefit from the drug ivacaftor based on mutations in the CFTR gene (5); and the poster child of personalised medicine, trastuzumab, is indicated for patients with metastatic breast cancer overexpressing the human epidermal growth factor receptor-2 (HER-2) (6). These examples, amongst others, demonstrating how unique genetic or molecular perturbations in an individual can lead to tailored therapy, led to strategic initiatives to scale up precision medicine to replicate these successes for other diseases.
Unlike conventional medicine, precision medicine is highly data-intensive and requires health data flow from individual medical records into different research contexts—for instance clinical trials, genomic research, pharmacovigilance, epidemiological studies—and then back into a learning healthcare system for the research outcomes to be integrated into practice (7). Research is an integral component of precision medicine, which requires data collected during the course of clinical care to be applied in the study of real-world clinical outcomes to enhance generalisability of the interventions. The recognition of the value and potential of precision medicine has led to the development of initiatives to accelerate and support research by collecting vast amounts of clinical and biomedical data. For example, the All of Us (AoU) research program (8) in the United States (formerly known as the Precision Medicine Initiative) aims to gather data from at least one million consenting individuals in the form of electronic health records (EHRs), biomarker and genomic analyses of donated tissue samples, mobile health devices and surveys. Similar initiatives have sprung up in both the public and private sectors across the world: 100,000 Genomes Project and UK Biobank in the UK; BioBank Japan; China Kadoorie Biobank; Biobank Graz in Austria; and FinnGen in Finland (9-12). These repositories of observational data are critical to the delivery of precision medicine despite the known limitations of observational studies to infer causality. This is described below, and some of the justifications for turning to observational data include the relative ease to collect large datasets, the difficulties in setting up randomised controlled trials (RCTs) for rare diseases and the huge sample sizes required for pharmacogenomic studies. Equally important to big data are the artificial intelligence (AI) tools which enable the extraction of clinically meaningful insights from the data (Figure 1).
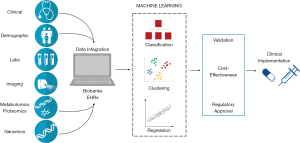
AI and Machine Learning (ML)
The terms AI and ML have been used interchangeably, but it is important to understand the differences between them. AI is a suite of technologies which enables a machine to simulate human behaviour. Two fields of AI which are particularly relevant to healthcare are ML and Natural Language Processing (NLP) (13,14). ML allows a software or algorithm to automatically learn from past data without programming explicitly, while NLP gives machines the ability to read, understand and derive meaning from human languages. We shall focus primarily on ML in this review, as most AI applications in healthcare are based on this subset of AI. The limitations of traditional statistical tools, such as linear and logistic regression which are commonly used for clinical outcome prediction, are well documented—they perform poorly with nonlinear relationships or high-dimensional data and fail to account for unknown interactions between input variables. Furthermore, in the era of big data, they are unnecessarily labour-intensive (15). In contrast to relatively rigid traditional statistical methods, the inherent flexibility of ML along with the scope for automation and the ability to learn from input data to progressively improve model performance without requiring explicit programming suggest ML is the right tool to be considered for precision medicine.
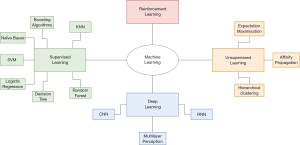
Whilst a detailed description of different ML algorithms is beyond the scope of this review, a summary of ML algorithms and their applications in healthcare research is presented in Table 1 (14). Although this is not an exhaustive list, it reflects the breadth of algorithms and clinical questions that are being explored using ML. ML can be categorized into four learning types: supervised, unsupervised, reinforcement and deep learning (Figure 2) (14). Supervised learning algorithms use a dataset labelled by humans to predict a specified or known outcome. Unsupervised learning algorithms, on the other hand, find patterns and associations in unlabelled data without human intervention. Supervised learning algorithms carry out classification and regression tasks, while unsupervised learning algorithms are limited to clustering. Reinforcement learning is a hybrid of supervised and unsupervised learning, which maximises the accuracy of algorithms using trial and error and thus is not applicable in the healthcare setting. Finally, deep learning, which is based on the structure of neural networks of the brain, is an autonomous system with multiple hidden layers of data processing. Deep learning algorithms independently find patterns even in unstructured data, which are then employed to make predictions about new data (111).
Table 1
| Clinical study | ML algorithm | Reference | |
|---|---|---|---|
| Rapid diagnosis of depression | Boosting algorithm | ( |
|
| Improving warfarin usage for the elderly inpatients | Boosting algorithm | ( |
|
| Predicting sepsis using vital sign data in the emergency department | Boosting algorithm | ( |
|
| Predicting adverse events in patients undergoing major cardiovascular procedures | Boosting algorithm | t( |
|
| Predicting urinary tract infections in the emergency department | Boosting algorithm | ( |
|
| Classifying lung nodules | Boosting algorithm | ( |
|
| Predicting transition from gestational diabetes mellitus to type 2 diabetes | Decision tree | ( |
|
| Identifying diffusion lesions in acute ischemic stroke | Decision tree | ( |
|
| Diabetic foot amputation risk analysis | Decision tree | ( |
|
| A top-down searching approach for diagnosis | Decision tree | ( |
|
| Performance surveillance of infant incubators | Decision tree | ( |
|
| Improving the prediction of total surgical procedure time | Decision tree | ( |
|
| Predicting graft survival for kidney transplantation | Ensemble methods | ( |
|
| Predicting treatment success in patients with substance use disorder | Ensemble methods | ( |
|
| Diagnosing breast cancer | Ensemble methods | ( |
|
| Detecting patients’ asthma control level | Ensemble methods | ( |
|
| Early prediction of outcome of cognitive behavioural therapy | Ensemble methods | ( |
|
| Automatic fall detection system for real-life monitoring | Hidden markov | ( |
|
| Detecting QRS complexes in single-lead ECG recordings | Hidden markov | ( |
|
| Real-time circadian phase estimation | Hidden markov | ( |
|
| Multi-channel EEG based automatic epileptic seizure detection | Hidden markov | ( |
|
| Real-time calibration and automatic drug dosing recommendation for chemotherapy treatment plans (Curate.AI) | Hidden markov | ( |
|
| Classifying prognostic phenotypes in heart failure patients | Hierarchical clustering | ( |
|
| Detecting disease-specific clusters within aortic arch images | Hierarchical clustering | ( |
|
| Clustering blood results in paediatric inflammatory bowel disease | Hierarchical clustering | ( |
|
| Predicting therapeutic response in IgG4-related disease | Hierarchical clustering | ( |
|
| Detecting thyroid diseases | Hierarchical clustering | ( |
|
| Classifying venomous and non-venomous snake bites | KNN | ( |
|
| Analysing and identifying kidney stone | KNN | ( |
|
| Predicting retinopathy risk | KNN | ( |
|
| Classifying lower back pain | KNN | ( |
|
| Distinguishing physiological from pathological patterns of hypertrophic remodelling | KNN | ( |
|
| Diagnosis of coronary artery disease | LDA | ( |
|
| Differentiating basal cell carcinoma and healthy skin | LDA | ( |
|
| Early diagnosis of mild cognitive impairment in Alzheimer's disease | LDA | ( |
|
| Predicting and classifying risk level of breast cancer | LDA | ( |
|
| Detecting and classifying dementia subtypes | LDA | ( |
|
| Identifying distinct bronchiectasis phenotypes | LDA | ( |
|
| Monitoring physician prescribing patterns and ensure the appropriateness of treatment | Linear regression | ( |
|
| Predicting conversion time to Alzheimer’s disease | Linear regression | ( |
|
| Predicting hypoxemia and Covid-19 disease outcome based on nasopharyngeal viral load | Linear regression | ( |
|
| Predicting maternal vitamin D status during pregnancy and lactation | Linear regression | ( |
|
| Predicting hospitalization and outpatient corticosteroid use in inflammatory bowel disease patients | Linear regression | ( |
|
| Differentiating severe septic patients with acute respiratory distress syndrome from those without | Logistic regression | ( |
|
| Early identification of patients with acute decompensated heart failure | Logistic regression | ( |
|
| Predicting early- and long-term mortality in hospitalized patients at risk of malnutrition | Logistic regression | ( |
|
| Predicting chemotherapy and radiotherapy outcome | Logistic regression | ( |
|
| Predicting autism spectrum disorder diagnosis | Logistic regression | ( |
|
| Microprocessor-based device for real-time prediction of acute cardiovascular events | Naïve Bayes | ( |
|
| Predicting atherosclerosis progression from ultrasound images | Naïve Bayes | ( |
|
| Detecting clinically important colorectal surgical site infection | Naïve Bayes | ( |
|
| Improving detection and diagnosis of bone tumour | Naïve Bayes | ( |
|
| Perceptron multilayer for classifying the risk in paediatric congenital heart surgery | Naïve Bayes | ( |
|
| Detecting diabetic retinopathy | Neural network | ( |
|
| Classifying skin cancer | Neural network | ( |
|
| Estimating optimal dose for intensity-modulated radiation therapy in prostate cancer patients | Neural network | ( |
|
| Identifying autism spectrum disorder from the brain images | Neural network | ( |
|
| Automatic cardiac arrhythmia detection on ECG | Neural network | ( |
|
| Identifying a molecular network predictive of advanced coronary calcium | Neural network | ( |
|
| Predicting knee osteoarthritis risk | Neural network | ( |
|
| Predicting adverse drug reactions and identifying the responsible molecular substructures | Neural network | ( |
|
| Predicting lower intestinal bleeding and need for surgical intervention | Neural network | ( |
|
| Automated classification of skin lesions using images | Neural network | ( |
|
| Automated detection of ischemic stroke | Neural network | ( |
|
| Chronic obstructive pulmonary disease staging and acute respiratory disease prediction in smokers | Neural network | ( |
|
| Risk assessment for major complications and death after surgery | Neural network | ( |
|
| Predicting early graft rejection in antibody incompatible kidney transplantation | Neural network | ( |
|
| Reinforcement learning for blood pressure regulation in post-cardiac surgery patients | Proprietary algorithms | ( |
|
| Predicting medical adherence of patients with heart failure | Proprietary algorithms | ( |
|
| Automatic IMRT planning in Philips Radiation Oncology Systems for head and neck cancer treatment | Proprietary algorithms | ( |
|
| Automated speech analysis to measure and predict psychosis onset | Proprietary algorithms | ( |
|
| The Seattle Heart Failure Model for heart failure survival analysis | Proprietary algorithms | ( |
|
| Predicting future myopia development in school children | Random forest | ( |
|
| Differentiating pituitary metastasis from autoimmune hypophysitis | Random forest | ( |
|
| Predicting early graft rejection in antibody incompatible kidney transplantation. | Random forest | ( |
|
| Predicting hospitalization and outpatient corticosteroid use in inflammatory bowel disease patients | Random forest | ( |
|
| Predicting rheumatoid arthritis mortality | Random forest | ( |
|
| Predicting in-hospital length of stay among cardiac patients | Random forest | ( |
|
| Predicting presence of advanced coronary artery calcium | Random forest | ( |
|
| Predicting risk of suicide attempts over time | Random forest | ( |
|
| Assessing risk of fibrosis and other liver-related outcomes in chronic Hepatitis C patients | Random forest | ( |
|
| Predicting long-term cognitive outcome following breast cancer | Random forest | ( |
|
| Predicting readmission rate in heart failure patients | Random forest | ( |
|
| Automatic detection of seizures in single-channel intra-cranial electroencephalograph recording | SVM | ( |
|
| Detecting structural imaging signature of schizophrenia | SVM | ( |
|
| Differentiating responders and non-responders to depression treatment | SVM | ( |
|
| Discriminating between hypovolemia and euvolemia using photoplethysmographic signals | SVM | ( |
|
| Predicting medication nonadherence in Crohn’s disease maintenance therapy | SVM | ( |
|
| Landmark text mining example of disease-chemical relationships to predict a benefit for using fish oil in Raynaud’s syndrome | Text mining | ( |
|
| Information retrieval and document triage in the Pharmacogenomics Database | Text mining | ( |
|
| Rule-based text mining approach for microRNA expression in cancer cells | Text mining | ( |
|
| Phenotype extraction from electronic health records | Text mining | ( |
|
| Patient outcome prediction through similarity analytics | Text mining | ( |
|
KNN, K-nearest neighbours; LDA, linear discriminant analysis; SVM, support vector machine.
NLP overlaps with ML (deep learning in particular) and has gained traction as a tool for data extraction from EHRs. A significant proportion of medical data contained in EHRs, such as descriptions of clinical features and diagnoses, is unstructured free text and the value of NLP is recognised for parsing clinical notes into practical data inputs including risk assessments (14,112). Furthermore, applying NLP to scientific literature has highlighted its potential for drug repurposing associations (113). In addition to these types of data mining, NLP may also have a future role in facilitating and automating patient engagement through chatbots. Chatbots simulate human conversations (in both written and spoken form) and are already widely used by online customer support services or virtual assistants such as Amazon Alexa. In healthcare, chatbots could serve as a stand-in for a physician as the first port of call to advise on symptoms and give preliminary diagnoses, freeing up some of the time of healthcare workers to focus on tasks which cannot be automated (114).
Prospects for AI in precision medicine
Success in implementing precision medicine in healthcare requires the communication and participation of people across a wide spectrum of disciplines including molecular biology, genetics, pathology, informatics, computer science, statistics and clinical science along with health economists, health insurers and hospital managers. The potential roles of AI span data-integration, making work-flows efficient and error-proof, generating clinically meaningful insights from big data, and developing new medicines. However, the published evidence across all these domains are predominantly represented by early-phase in silico studies with little validation and do not fully cover potential sources of bias specific to AI systems (115). In relation to precision medicine, ML approaches mainly feature in three areas with tangible successes evident in the first two (116): (I) prediction of pharmaceutical properties of molecular compounds and targets for drug discovery (117); (II) faster diagnosis using pattern recognition and segmentation techniques on medical images (from, e.g., retinal scans, pathology slides and body surfaces, bones and internal organs) (71,72,118); and (III) the development of predictive models using deep learning techniques on multimodal data sources such as genomic and clinical data (119). The paucity of use cases of AI in precision medicine may partly be related to studies not conforming to RCT level rigour that is crucial for regulatory approval and clinical adoption.
Technical challenges of ML in precision medicine
Despite technical advancements in informatics, computer science and mathematics, the development and application of ML models remains a challenging process. When building ML models, data is split into training and testing sets. The training set teaches the model, while the model’s performance is evaluated by how well it describes the testing set. Researchers typically split the data at random. However, data in real life are rarely random and show trends over time, for instance differences data collection processes, measurement methods or changes in guidelines. These variations can have an impact on prediction accuracies.
Overfitting and underfitting
Overfitting is a major issue for all ML models. Overfitting arises when an algorithm learns to make rules which fit both random noise and meaningful signals in training data accurately and specifically, but fail with testing data (120). The resulting algorithm can thus perfectly predict the training data but only at the price of its performance on new data. Because of the availability of an enormous variety of clinical variables recorded in EHRs and biobanks, it can be tempting to develop a highly specific model with a multitude of predictive features for a given disease. Such a model is likely to show excellent performance in its training dataset but fail in validation, limiting applicability in the real world, resulting in little more than wasted healthcare resources. This is the opposite of underfitting, which occurs when a model is too simple (informed by too few features or regularized too much) making it inflexible in learning from the dataset (121). Overfitting and underfitting can be overcome by making modifications to the training set or by optimising the parameters of the model. For these reasons, training of a good model often requires large datasets, competent informatics skills, domain knowledge and an adequate means of validation (121). When a model is developed by an ML expert with little knowledge of the clinical context of variables within an EHR, feature selection may be misguided leading to erroneous predictions and poor model performance. Thus, collaboration between medical and informatics experts is essential to extract value from big data.
Interpretability and explainability
As ML (especially deep learning) models grow more sophisticated and powerful with multiple hidden layers of data transformation and analysis between the input and output data, their decision-making process becomes more challenging for a human to conceptualise. While interpretability implies some sense of understanding how the technology works, explainability implies that a wider range of users can understand why or how a conclusion was reached. The complex architecture of deep learning models, for example, makes them more difficult to understand and interpret than their supervised and unsupervised counterparts (111). Lack of interpretability presents one of the greatest barriers to the acceptance of ML into clinical practice. For instance, if a clinical expert and an ML algorithm are presented with the same patient data but arrive at different diagnoses, it may not be possible to interrogate the algorithm to find where and why the decision-making processes diverge. The translation of these so-called “black-box models” into clinical practice thus requires institutions and researchers to place a significant amount of blind trust in the development process of the algorithms, as well as in the soundness of the data on which the models were trained, tested and validated. This is particularly problematic in clinical practice given the potentially devastating and long-lasting consequences of a faulty model to patients’ wellbeing (122).
Each model is unique and often requires a combination of mathematical equations, verbal descriptions and visual representations, such as Bayesian networks, to communicate and justify their inner workings (123). The development of interpretable models will not only improve trust in their logic but will also allow the users of the model to identify faulty or infeasible outputs. Furthermore, the ability to interpret and question a model’s behaviour may reveal patterns and associations in data which would not be recognised by humans, providing valuable scientific insight. The importance of this is reflected in the considerable amount of research being carried out in the field of explainable AI (XAI) (124).
Observational data and causality
While it is clear that ML can be used to discover previously unknown associations in observational data, it is often forgotten that observational data cannot be used to directly infer causality. Establishing causality requires not only sufficient information about the environment of measured variables, but also the removal of various biases that undermine observational studies (125). RCTs meet both of these conditions and are thus considered the gold standard for deducing causality (126). Observational data, on the other hand, rarely contain enough contextual information to allow for robust causal data analysis. This is particularly problematic if correlations are mistaken for causation when developing health intervention models, which have a direct impact on clinical care (126). As most of the big data resources that are available for ML are cross-sectional observational data, rather than controlled clinical trials, solutions are needed to overcome these limitations. One strategy to infer causality from observational data is to model counterfactual predictions. Counterfactuals allow us to ask “What would have happened, had a different intervention been applied?”, rather than being limited to “What happened?” or “What will happen?” and this is a line of research that shows promise (123).
Algorithmic bias
Algorithmic unfairness poses another major obstacle to the translation of ML to clinical practice. Because ML models adaptively improve their performances by “learning” information directly from the data, the success of the model is acutely sensitive to data quality. Under-representation of minority subgroups in the dataset may create a blind spot in the model and introduce a discriminatory bias toward that group of patients (127). The problem is almost always an unintended property of the dataset and not the conscious choice of the researchers, but there is a risk of the model being implemented into practice with the algorithmic bias remaining undetected. For instance, multiple patients from African or unspecified ancestry had their benign genetic variants misclassified as pathogenic (128). The cause for genetic misdiagnoses and potential health disparities was identified as the failure to account for genetic diversity in non-European populations at the time of testing. The misclassification was resolved with the inclusion of genetic data for African ethnic patients in the training groups (128). For this reason, algorithms should always be designed after careful consideration of all relevant variations in patient demographics and pathologic states in real-world settings. This is to ensure the training data truly represents the population of the intended deployment community.
Validation and generalisability
Other than technical limitations, there are other issues that obstruct the realisation of ML’s potential in real-world practices. Medicine is an ever-changing field, where clinical and operational practices in clinical settings constantly evolve. The introduction of an ML algorithm may lead to a paradigm shift in normal practice. Subsequently, the input data will also change and no longer resemble the data that was used to train the model (129). To maintain performance over time, models should be constantly monitored for deteriorating performance and may need to be subjected to periodical recalibration or retraining. Dataset shift may occur due to the technical differences between institutions as well as variations in local clinical practices (130). Thus, it can be challenging to implement an ML model in a different setting to where it was originally trained. This issue can be mitigated by conducting site-specific training to adapt the existing model to a new study population. For instance, a deep learning model for diagnosing diabetic retinopathy from medical images failed to perform as expected when implemented across clinics in Thailand, partly due to the fact that the model was trained with images collected from clinics with different lighting conditions (131).
For all these reasons, the generalisability and real-world performance of the model need to be externally validated against adequately sized datasets from institutions other than where it was originally trained. However, there is a critical scarcity of public healthcare repositories at the present time. Multitudes of usage policies, security, and privacy concerns further add to the complexity of data collaboration (132). A 2019 systematic review for the diagnostic use of ML in medical imaging found that external validation was performed only on 6% of 516 published studies (133).
Finally, ML models, although not being a major concern at present, are susceptible to adversarial attack and manipulation (134). We must always be mindful that machine learning models do not truly learn and understand their tasks – at least not in the same way that a human does. The models are simply a chain of mathematical algorithms, designed to mathematically map the input data to the targets. As such, ML models are brittle and can be easily fooled by explicitly designed inputs (134). For example, by adding the noise from a picture of a malignant mole to that of a benign mole, a study managed to trick the ML model into classifying the benign mole as malignant (135).
These findings highlight an alarming trend that AI studies are neither carried out nor reported with the same rigour as other medical research. To improve the quality of scientific AI investigations and to accelerate the clinical adoption of AI-based solutions, AI extensions have been added to both the SPIRIT (Standard Protocol Items: Recommendations for Interventional Trials) and CONSORT (Consolidated Standards of Reporting Trials) guidelines (SPIRIT-AI and CONSORT-AI, respectively) (136).
Conclusions
The realisation of precision medicine requires effective distillation of high-dimensional data across clinical, biological, patient-generated and environmental domains. AI, especially ML, is a critical enabler in this respect. The low-hanging fruit of ML in medicine are already successfully deployed in automating routine clinical processes to reduce the burden on clinical staff, for instance by prioritizing triage order in the emergency department or automating medical image evaluation. Whilst most of ML is currently based on analysis of observational data, which limits causal inference, the convergence of computational power, data, algorithms and greater traction with healthcare researchers has created the right inflection point for an exponential growth in rigorous discovery and implementation studies in precision medicine, as reflected in the number of publications in the area over the last five years. The hype hitherto associated with the precision medicine and ML narrative has waned with the growing realisation that ML is not a quick fix obviating the need for clinical and scientific expertise and scrutiny. However, to ensure that these applications are clinically useful and operationally feasible, a number of well-recognised methodological challenges still need to be overcome along with development of a rigorous framework for evidence generation demonstrating patient safety and benefit fulfilling all regulatory requirements.
Acknowledgments
Funding: SP and CdT acknowledge support from Health Data Research UK which receives its funding from HDR UK Ltd (HDR-5012) funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation (BHF) and the Wellcome Trust.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Hospital Management and Health Policy for the series “AI in Healthcare - Opportunities and Challenges”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jhmhp-20-132). The series “AI in Healthcare - Opportunities and Challenges” was commissioned by the editorial office without any funding or sponsorship. Sandosh Padmanabhan served as the unpaid Guest Editor of the series. Clea du Toit and Sandosh Padmanabhan acknowledge support from Health Data Research UK which receives its funding from HDR UK Ltd (HDR-5012) funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation (BHF) and the Wellcome Trust. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793-5. [Crossref] [PubMed]
- Schork NJ. Personalized medicine: Time for one-person trials. Nature 2015;520:609-11. [Crossref] [PubMed]
- Goldberger JJ, Buxton AE. Personalized medicine vs guideline-based medicine. JAMA 2013;309:2559-60. [Crossref] [PubMed]
- Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med 1996;2:561-6. [Crossref] [PubMed]
- Davis PB, Yasothan U, Kirkpatrick P. Ivacaftor. Nat Rev Drug Discov 2012;11:349-50. [Crossref] [PubMed]
- Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002;20:719-26. [Crossref] [PubMed]
- Beauvais M, Knoppers BM. When information is the treatment? Precision medicine in healthcare. Healthc Manage Forum 2020;33:120-5. [Crossref] [PubMed]
- All of Us Research Program Investigators. The "All of Us" Research Program. N Engl J Med 2019;381:668-76. [Crossref] [PubMed]
- Chen Z, Chen J, Collins R, et al. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol 2011;40:1652-66. [Crossref] [PubMed]
- Sakaue S, Kanai M, Karjalainen J, et al. Trans-biobank analysis with 676,000 individuals elucidates the association of polygenic risk scores of complex traits with human lifespan. Nat Med 2020;26:542-8. [Crossref] [PubMed]
- Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779 [Crossref] [PubMed]
- Shilo S, Rossman H, Segal E. Axes of a revolution: challenges and promises of big data in healthcare. Nat Med 2020;26:29-38. [Crossref] [PubMed]
- Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc 2011;18:544-51. [Crossref] [PubMed]
- Ray S. editor. A Quick Review of Machine Learning Algorithms. 2019 International Conference on Machine Learning, Big Data, Cloud and Parallel Computing (COMITCon); 2019.
- Krittanawong C, Zhang H, Wang Z, et al. Artificial Intelligence in Precision Cardiovascular Medicine. J Am Coll Cardiol 2017;69:2657-64. [Crossref] [PubMed]
- Sharma A, Verbeke WJMI. Improving Diagnosis of Depression With XGBOOST Machine Learning Model and a Large Biomarkers Dutch Dataset (n = 11,081). Front Big Data 2020;3:15. [Crossref] [PubMed]
- Liu KE, Lo CL, Hu YH. Improvement of adequate use of warfarin for the elderly using decision tree-based approaches. Methods Inf Med 2014;53:47-53. [Crossref] [PubMed]
- Mao Q, Jay M, Hoffman JL, et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open 2018;8:e017833 [Crossref] [PubMed]
- Mortazavi BJ, Desai N, Zhang J, et al. Prediction of Adverse Events in Patients Undergoing Major Cardiovascular Procedures. IEEE J Biomed Health Inform 2017;21:1719-29. [Crossref] [PubMed]
- Taylor RA, Moore CL, Cheung KH, et al. Predicting urinary tract infections in the emergency department with machine learning. PLoS One 2018;13:e0194085 [Crossref] [PubMed]
- Nishio M, Nishizawa M, Sugiyama O, et al. Computer-aided diagnosis of lung nodule using gradient tree boosting and Bayesian optimization. PLoS One 2018;13:e0195875 [Crossref] [PubMed]
- Allalou A, Nalla A, Prentice KJ, et al. A Predictive Metabolic Signature for the Transition From Gestational Diabetes Mellitus to Type 2 Diabetes. Diabetes 2016;65:2529-39. [Crossref] [PubMed]
- Boldsen JK, Engedal TS, Pedraza S, et al. Better Diffusion Segmentation in Acute Ischemic Stroke Through Automatic Tree Learning Anomaly Segmentation. Front Neuroinform 2018;12:21. [Crossref] [PubMed]
- Kasbekar PU, Goel P, Jadhav SP. A Decision Tree Analysis of Diabetic Foot Amputation Risk in Indian Patients. Front Endocrinol (Lausanne) 2017;8:25. [Crossref] [PubMed]
- Khan RS, Zardar AA, Bhatti Z. Artificial Intelligence based Smart Doctor using Decision Tree Algorithm. arXiv preprint arXiv:1808.01884, 2018.
- Kovačević Ž, Gurbeta Pokvić L, Spahić L, et al. Prediction of medical device performance using machine learning techniques: infant incubator case study. Health Technol 2020;10:151-5. [Crossref]
- Edelman ER, van Kuijk SMJ, Hamaekers AEW, et al. Improving the Prediction of Total Surgical Procedure Time Using Linear Regression Modeling. Front Med (Lausanne) 2017;4:85. [Crossref] [PubMed]
- Yoo KD, Noh J, Lee H, et al. A Machine Learning Approach Using Survival Statistics to Predict Graft Survival in Kidney Transplant Recipients: A Multicenter Cohort Study. Sci Rep 2017;7:8904. [Crossref] [PubMed]
- Topuz K, Zengul FD, Dag A, et al. Predicting graft survival among kidney transplant recipients: A Bayesian decision support model. Decis Support Syst 2018;106:97-109. [Crossref]
- Acion L, Kelmansky D, van der Laan M, et al. Use of a machine learning framework to predict substance use disorder treatment success. PLoS One 2017;12:e0175383 [Crossref] [PubMed]
- Wang H, Zheng B, Yoon SW, et al. A support vector machine-based ensemble algorithm for breast cancer diagnosis. Eur J Oper Res 2018;267:687-99. [Crossref]
- Khasha R, Sepehri MM, Mahdaviani SA. An ensemble learning method for asthma control level detection with leveraging medical knowledge-based classifier and supervised learning. J Med Syst 2019;43:158. [Crossref] [PubMed]
- Safinianaini N, Boström H, Kaldo V, editors. Gated Hidden Markov Models for Early Prediction of Outcome of Internet-Based Cognitive Behavioral Therapy. Artificial Intelligence in Medicine. Cham: Springer International Publishing. 2019.
- Yu S, Chen H, Brown RA. Hidden Markov Model-Based Fall Detection With Motion Sensor Orientation Calibration: A Case for Real-Life Home Monitoring. IEEE J Biomed Health Inform 2018;22:1847-53. [Crossref] [PubMed]
- Monroy NF, Altuve M, editors. Analysis of the Observation Sequence Duration of Hidden Markov Models for QRS Complex Detection in Single-Lead ECG Recordings. 2018 Computing in Cardiology Conference (CinC); 2018.
- Komarzynski S, Bolborea M, Huang Q, et al. Predictability of individual circadian phase during daily routine for medical applications of circadian clocks. JCI Insight 2019;4:e130423 [Crossref] [PubMed]
- Dash DP, Kolekar MH, Jha K. Multi-channel EEG based automatic epileptic seizure detection using iterative filtering decomposition and Hidden Markov Model. Comput Biol Med 2020;116:103571 [Crossref] [PubMed]
- Zarrinpar A, Lee DK, Silva A, et al. Individualizing liver transplant immunosuppression using a phenotypic personalized medicine platform. Sci Transl Med 2016;8:333ra49 [Crossref] [PubMed]
- Pantuck AJ, Lee DK, Kee T, et al. Modulating BET Bromodomain Inhibitor ZEN-3694 and Enzalutamide Combination Dosing in a Metastatic Prostate Cancer Patient Using CURATE.AI, an Artificial Intelligence Platform. Adv Ther 2018;1:1800104 [Crossref]
- Przewlocka-Kosmala M, Marwick TH, Dabrowski A, et al. Contribution of Cardiovascular Reserve to Prognostic Categories of Heart Failure With Preserved Ejection Fraction: A Classification Based on Machine Learning. J Am Soc Echocardiogr 2019;32:604-15.e6. [Crossref] [PubMed]
- Bruse JL, Zuluaga MA, Khushnood A, et al. Detecting Clinically Meaningful Shape Clusters in Medical Image Data: Metrics Analysis for Hierarchical Clustering Applied to Healthy and Pathological Aortic Arches. IEEE Trans Biomed Eng 2017;64:2373-83. [Crossref] [PubMed]
- Ashton JJ, Borca F, Mossotto E, et al. Analysis and Hierarchical Clustering of Blood Results Before Diagnosis in Pediatric Inflammatory Bowel Disease. Inflamm Bowel Dis 2020;26:469-75. [PubMed]
- Yamamoto M, Takano KI, Kamekura R, et al. Predicting therapeutic response in IgG4-related disease based on cluster analysis. Immunol Med 2018;41:30-3. [Crossref] [PubMed]
- Chandel K, Kunwar V, Sabitha S, et al. A comparative study on thyroid disease detection using K-nearest neighbor and Naive Bayes classification techniques. CSI Transactions on ICT 2016;4:313-9. [Crossref]
- Putra RM. Adiwijaya, Utama DQ. Snake bite classification using Chain code and K nearest neighbour. Journal of Physics: Conference Series 2019;1192:012015 [Crossref]
- Verma J, Nath M, Tripathi P, et al. Analysis and identification of kidney stone using Kth nearest neighbour (KNN) and support vector machine (SVM) classification techniques. Pattern Recognition and Image Analysis 2017;27:574-80. [Crossref]
- Arun V, Prajwal V, Girish A, et al. editors. Diabetic Retinopathy Risk Prediction for Diabetics Using Nearest Neighbour Approach. Emerging Research in Electronics, Computer Science and Technology. Singapore: Springer Singapore, 2019.
- Sandag GA, Tedry NE, Lolong S, editors. Classification of Lower Back Pain Using K-Nearest Neighbor Algorithm. 2018 6th International Conference on Cyber and IT Service Management (CITSM); 2018.
- Narula S, Shameer K, Salem Omar AM, et al. Machine-Learning Algorithms to Automate Morphological and Functional Assessments in 2D Echocardiography. J Am Coll Cardiol 2016;68:2287-95. [Crossref] [PubMed]
- Kolukisa B, Hacilar H, Goy G, et al. editors. Evaluation of Classification Algorithms, Linear Discriminant Analysis and a New Hybrid Feature Selection Methodology for the Diagnosis of Coronary Artery Disease. 2018 IEEE International Conference on Big Data (Big Data); 2018.
- Chernomyrdin N, Zaytsev K, Lesnichaya A, et al. Principle component analysis and linear discriminant analysis of multi-spectral autofluorescence imaging data for differentiating basal cell carcinoma and healthy skin. SPIE Optical Engineering + Applications. SPIE; 2016.
- Fang C, Li C, Cabrerizo M, et al. editors. A Gaussian discriminant analysis-based generative learning algorithm for the early diagnosis of mild cognitive impairment in Alzheimer's disease. 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2017.
- Rajaguru H, Prabhakar SK. editors. Bayesian linear discriminant analysis for breast cancer classification. 2017 2nd International Conference on Communication and Electronics Systems (ICCES); 2017.
- Neto E, Biessmann F, Aurlien H, et al. Regularized Linear Discriminant Analysis of EEG Features in Dementia Patients. Front Aging Neurosci 2016;8:273. [Crossref] [PubMed]
- Guan WJ, Jiang M, Gao YH, et al. Unsupervised learning technique identifies bronchiectasis phenotypes with distinct clinical characteristics. Int J Tuberc Lung Dis 2016;20:402-10. [Crossref] [PubMed]
- Backenroth D, Chase HS, Wei Y, et al. Monitoring prescribing patterns using regression and electronic health records. BMC Med Inform Decis Mak 2017;17:175. [Crossref] [PubMed]
- Yang SJ, Shin H, Lee SH, et al. Functional linear regression model with randomly censored data: Predicting conversion time to Alzheimer ’s disease. Comput Stat Data Anal 2020;150:107009 [Crossref]
- Shlomai A, Ben-Zvi H, Glusman Bendersky A, et al. Nasopharyngeal viral load predicts hypoxemia and disease outcome in admitted COVID-19 patients. Crit Care 2020;24:539. [Crossref] [PubMed]
- Wagner C, Park Y, Shary J, et al. Mathematical Models to Predict Maternal Vitamin D (VitD) Status During Pregnancy and Lactation. Curr Dev Nutr 2020;4:1847. [Crossref]
- Waljee AK, Lipson R, Wiitala WL, et al. Predicting Hospitalization and Outpatient Corticosteroid Use in Inflammatory Bowel Disease Patients Using Machine Learning. Inflamm Bowel Dis 2017;24:45-53. [Crossref] [PubMed]
- Ware LB, Koyama T, Zhao Z, et al. Biomarkers of lung epithelial injury and inflammation distinguish severe sepsis patients with acute respiratory distress syndrome. Crit Care 2013;17:R253. [Crossref] [PubMed]
- Blecker S, Sontag D, Horwitz LI, et al. Early Identification of Patients With Acute Decompensated Heart Failure. J Card Fail 2018;24:357-62. [Crossref] [PubMed]
- Sanson G, Sadiraj M, Barbin I, et al. Prediction of early- and long-term mortality in adult patients acutely admitted to internal medicine: NRS-2002 and beyond. Clin Nutr 2020;39:1092-100. [Crossref] [PubMed]
- Deist TM, Dankers F, Valdes G, et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers. Med Phys 2018;45:3449-59. [Crossref] [PubMed]
- Altay O, Ulas M. editors. Prediction of the autism spectrum disorder diagnosis with linear discriminant analysis classifier and K-nearest neighbor in children. 2018 6th International Symposium on Digital Forensic and Security (ISDFS); 2018 22-25.
- Tylman W, Waszyrowski T, Napieralski A, et al. Real-time prediction of acute cardiovascular events using hardware-implemented Bayesian networks. Comput Biol Med 2016;69:245-53. [Crossref] [PubMed]
- Hu X, Reaven PD, Saremi A, et al. Machine learning to predict rapid progression of carotid atherosclerosis in patients with impaired glucose tolerance. EURASIP J Bioinform Syst Biol 2016;2016:14. [Crossref] [PubMed]
- Sohn S, Larson DW, Habermann EB, et al. Detection of clinically important colorectal surgical site infection using Bayesian network. J Surg Res 2017;209:168-73. [Crossref] [PubMed]
- Do BH, Langlotz C, Beaulieu CF. Bone Tumor Diagnosis Using a Naive Bayesian Model of Demographic and Radiographic Features. J Digit Imaging 2017;30:640-7. [Crossref] [PubMed]
- Ruiz-Fernández D, Monsalve Torra A, Soriano-Payá A, et al. Aid decision algorithms to estimate the risk in congenital heart surgery. Comput Methods Programs Biomed 2016;126:118-27. [Crossref] [PubMed]
- Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016;316:2402-10. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Nguyen D, Long T, Jia X, et al. A feasibility study for predicting optimal radiation therapy dose distributions of prostate cancer patients from patient anatomy using deep learning. Sci Rep 2019;9:1076. [Crossref] [PubMed]
- Heinsfeld AS, Franco AR, Craddock RC, et al. Identification of autism spectrum disorder using deep learning and the ABIDE dataset. Neuroimage Clin 2017;17:16-23. [Crossref] [PubMed]
- Parvaneh S, Rubin J, Babaeizadeh S, et al. Cardiac arrhythmia detection using deep learning: A review. J Electrocardiol 2019;57S:S70-4. [Crossref] [PubMed]
- Tran L, Li Y, Nocera L, et al. MultiFusionNet: Atrial Fibrillation Detection With Deep Neural Networks. AMIA Jt Summits Transl Sci Proc 2020;2020:654-63. [PubMed]
- Oguz C, Sen SK, Davis AR, et al. Genotype-driven identification of a molecular network predictive of advanced coronary calcium in ClinSeq(R) and Framingham Heart Study cohorts. BMC Syst Biol 2017;11:99. [Crossref] [PubMed]
- Yoo TK, Kim DW, Choi SB, et al. Simple Scoring System and Artificial Neural Network for Knee Osteoarthritis Risk Prediction: A Cross-Sectional Study. PLoS One 2016;11:e0148724 [Crossref] [PubMed]
- Dey S, Luo H, Fokoue A, et al. Predicting adverse drug reactions through interpretable deep learning framework. BMC Bioinformatics 2018;19:476. [Crossref] [PubMed]
- Loftus TJ, Brakenridge SC, Croft CA, et al. Neural network prediction of severe lower intestinal bleeding and the need for surgical intervention. J Surg Res 2017;212:42-7. [Crossref] [PubMed]
- Murray NM, Unberath M, Hager GD, et al. Artificial intelligence to diagnose ischemic stroke and identify large vessel occlusions: a systematic review. J Neurointerv Surg 2020;12:156-64. [Crossref] [PubMed]
- González G, Ash SY, Vegas-Sánchez-Ferrero G, et al. Disease Staging and Prognosis in Smokers Using Deep Learning in Chest Computed Tomography. Am J Respir Crit Care Med 2018;197:193-203. [Crossref] [PubMed]
- Bihorac A, Ozrazgat-Baslanti T, Ebadi A, et al. MySurgeryRisk: Development and Validation of a Machine-learning Risk Algorithm for Major Complications and Death After Surgery. Ann Surg 2019;269:652-62. [Crossref] [PubMed]
- Shaikhina T, Lowe D, Daga S, et al. Decision tree and random forest models for outcome prediction in antibody incompatible kidney transplantation. Biomed Signal Process Control 2019;52:456-62. [Crossref]
- Sandu C, Popescu D, Popescu C. editors. Post cardiac surgery recovery process with reinforcement learning. 2015 19th International Conference on System Theory, Control and Computing (ICSTCC); 2015.
- Son YJ, Kim HG, Kim EH, et al. Application of support vector machine for prediction of medication adherence in heart failure patients. Healthc Inform Res 2010;16:253-9. [Crossref] [PubMed]
- Karanasiou GS, Tripoliti EE, Papadopoulos TG, et al. Predicting adherence of patients with HF through machine learning techniques. Healthc Technol Lett 2016;3:165-70. [Crossref] [PubMed]
- Hazell I, Bzdusek K, Kumar P, et al. Automatic planning of head and neck treatment plans. J Appl Clin Med Phys 2016;17:272-82. [Crossref] [PubMed]
- Kusters JMAM, Bzdusek K, Kumar P, et al. Automated IMRT planning in Pinnacle: A study in head-and-neck cancer. Strahlenther Onkol 2017;193:1031-8. [Crossref] [PubMed]
- Bedi G, Carrillo F, Cecchi GA, et al. Automated analysis of free speech predicts psychosis onset in high-risk youths. NPJ Schizophr 2015;1:15030. [Crossref] [PubMed]
- Panahiazar M, Taslimitehrani V, Pereira N, et al. Using EHRs and Machine Learning for Heart Failure Survival Analysis. Stud Health Technol Inform 2015;216:40-4. [PubMed]
- Lin H, Long E, Ding X, et al. Prediction of myopia development among Chinese school-aged children using refraction data from electronic medical records: A retrospective, multicentre machine learning study. PLoS Med 2018;15:e1002674 [Crossref] [PubMed]
- Mekki A, Dercle L, Lichtenstein P, et al. Machine learning defined diagnostic criteria for differentiating pituitary metastasis from autoimmune hypophysitis in patients undergoing immune checkpoint blockade therapy. Eur J Cancer 2019;119:44-56. [Crossref] [PubMed]
- Lezcano-Valverde JM, Salazar F, Leon L, et al. Development and validation of a multivariate predictive model for rheumatoid arthritis mortality using a machine learning approach. Sci Rep 2017;7:10189. [Crossref] [PubMed]
- Daghistani TA, Elshawi R, Sakr S, et al. Predictors of in-hospital length of stay among cardiac patients: A machine learning approach. Int J Cardiol 2019;288:140-7. [Crossref] [PubMed]
- Walsh CG, Ribeiro JD, Franklin JC. Predicting Risk of Suicide Attempts Over Time Through Machine Learning. Clin Psychol Sci 2017;5:457-69. [Crossref]
- Konerman MA, Lu D, Zhang Y, et al. Assessing risk of fibrosis progression and liver-related clinical outcomes among patients with both early stage and advanced chronic hepatitis C. PLoS One 2017;12:e0187344 [Crossref] [PubMed]
- Kesler SR, Rao A, Blayney DW, et al. Predicting Long-Term Cognitive Outcome Following Breast Cancer with Pre-Treatment Resting State fMRI and Random Forest Machine Learning. Front Hum Neurosci 2017;11:555. [Crossref] [PubMed]
- Shameer K, Johnson KW, Yahi A, et al. Predictive Modeling of Hospital Readmission Rates Using Electronic Medical Record-Wide Machine Learning: A Case-Study Using Mount Sinai Heart Failure Cohort. Pac Symp Biocomput 2017;22:276-87. [Crossref] [PubMed]
- Zhang Zisheng, Parhi KK. Seizure detection using wavelet decomposition of the prediction error signal from a single channel of intra-cranial EEG. Annu Int Conf IEEE Eng Med Biol Soc 2014;2014:4443-6. [Crossref] [PubMed]
- Rozycki M, Satterthwaite TD, Koutsouleris N, et al. Multisite Machine Learning Analysis Provides a Robust Structural Imaging Signature of Schizophrenia Detectable Across Diverse Patient Populations and Within Individuals. Schizophr Bull 2018;44:1035-44. [Crossref] [PubMed]
- Bailey NW, Hoy KE, Rogasch NC, et al. Differentiating responders and non-responders to rTMS treatment for depression after one week using resting EEG connectivity measures. J Affect Disord 2019;242:68-79. [Crossref] [PubMed]
- Reljin N, Zimmer G, Malyuta Y, et al. Using support vector machines on photoplethysmographic signals to discriminate between hypovolemia and euvolemia. PLoS One 2018;13:e0195087 [Crossref] [PubMed]
- Wang L, Fan R, Zhang C, et al. Applying Machine Learning Models to Predict Medication Nonadherence in Crohn's Disease Maintenance Therapy. Patient Prefer Adherence 2020;14:917-26. [Crossref] [PubMed]
- Swanson DR. Fish oil, Raynaud's syndrome, and undiscovered public knowledge. Perspect Biol Med 1986;30:7-18. [Crossref] [PubMed]
- Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics 2010;11:501-5. [Crossref] [PubMed]
- Xie B, Ding Q, Han H, et al. miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics 2013;29:638-44. [Crossref] [PubMed]
- Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc 2015;22:993-1000. [Crossref] [PubMed]
- Liao KP, Sun J, Cai TA, et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J Am Med Inform Assoc 2019;26:1255-62. [Crossref] [PubMed]
- Lee J, Maslove DM, Dubin JA. Personalized mortality prediction driven by electronic medical data and a patient similarity metric. PLoS One 2015;10:e0127428 [Crossref] [PubMed]
- Zhang S, Bamakan SMH, Qu Q, et al. Learning for Personalized Medicine: A Comprehensive Review From a Deep Learning Perspective. IEEE Rev Biomed Eng 2019;12:194-208. [Crossref] [PubMed]
- Feller DJ, Zucker J, Yin MT, et al. Using Clinical Notes and Natural Language Processing for Automated HIV Risk Assessment. J Acquir Immune Defic Syndr 2018;77:160-6. [Crossref] [PubMed]
- Yang HT, Ju JH, Wong YT, et al. Literature-based discovery of new candidates for drug repurposing. Brief Bioinform 2017;18:488-97. [PubMed]
- Milne-Ives M, de Cock C, Lim E, et al. The Effectiveness of Artificial Intelligence Conversational Agents in Health Care: Systematic Review. J Med Internet Res 2020;22:e20346 [Crossref] [PubMed]
- Liu X, Faes L, Kale AU, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health 2019;1:e271-97. [Crossref] [PubMed]
- Shah P, Kendall F, Khozin S, et al. Artificial intelligence and machine learning in clinical development: a translational perspective. NPJ Digit Med 2019;2:69. [Crossref] [PubMed]
- Vamathevan J, Clark D, Czodrowski P, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov 2019;18:463-77. [Crossref] [PubMed]
- Rana A, Yauney G, Lowe A, et al. Computational Histological Staining and Destaining of Prostate Core Biopsy RGB Images with Generative Adversarial Neural Networks. 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA). IEEE, 2018.
- Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018;1:18. [Crossref] [PubMed]
- Mutasa S, Sun S, Ha R. Understanding artificial intelligence based radiology studies: What is overfitting? Clin Imaging 2020;65:96-9. [Crossref] [PubMed]
- Handelman GS, Kok HK, Chandra RV, et al. Peering Into the Black Box of Artificial Intelligence: Evaluation Metrics of Machine Learning Methods. AJR Am J Roentgenol 2019;212:38-43. [Crossref] [PubMed]
- . The Lancet Respiratory M. Opening the black box of machine learning. Lancet Respir Med 2018;6:801. [Crossref]
-
Pearl J. Theoretical Impediments to Machine Learning With Seven Sparks from the Causal Revolution. arXiv:180104016.2018 .10.1145/3159652.3176182 - Jiménez-Luna J, Grisoni F, Schneider G. Drug discovery with explainable artificial intelligence. Nat Mach Intell 2020;2:573-84. [Crossref]
- Grimes DA, Schulz KF. Bias and causal associations in observational research. Lancet 2002;359:248-52. [Crossref] [PubMed]
- Prosperi M, Guo Y, Sperrin M, et al. Causal inference and counterfactual prediction in machine learning for actionable healthcare. Nat Mach Intell 2020;2:369-75. [Crossref]
- Paulus JK, Kent DM. Predictably unequal: understanding and addressing concerns that algorithmic clinical prediction may increase health disparities. NPJ Digit Med 2020;3:99. [Crossref] [PubMed]
- Manrai AK, Funke BH, Rehm HL, et al. Genetic Misdiagnoses and the Potential for Health Disparities. N Engl J Med 2016;375:655-65. [Crossref] [PubMed]
- Nestor B, Mcdermott M, Chauhan G, et al. Rethinking clinical prediction: Why machine learning must consider year of care and feature aggregation. arXiv 2018;1811.12583.
- Kelly CJ, Karthikesalingam A, Suleyman M, et al. Key challenges for delivering clinical impact with artificial intelligence. BMC Med 2019;17:195. [Crossref] [PubMed]
- Beede E, Baylor E, Hersch F, et al. A Human-Centered Evaluation of a Deep Learning System Deployed in Clinics for the Detection of Diabetic Retinopathy. Proceedings of the 2020 CHI Conference on Human Factors in Computing Systems; Honolulu, HI, USA: Association for Computing Machinery; 2020:1-12.
- Schmitt CP, Burchinal M. Data management practices for collaborative research. Front Psychiatry 2011;2:47. [Crossref] [PubMed]
- Kim DW, Jang HY, Kim KW, et al. Design Characteristics of Studies Reporting the Performance of Artificial Intelligence Algorithms for Diagnostic Analysis of Medical Images: Results from Recently Published Papers. Korean J Radiol 2019;20:405-10. [Crossref] [PubMed]
- Heaven D. Why deep-learning AIs are so easy to fool. Nature 2019;574:163-6. [Crossref] [PubMed]
- Finlayson SG, Bowers JD, Ito J, et al. Adversarial attacks on medical machine learning. Science 2019;363:1287-9. [Crossref] [PubMed]
- Liu X, Cruz Rivera S, Moher D, et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nature Medicine 2020;26:1364-74. [Crossref] [PubMed]
Cite this article as: Tran TQB, du Toit C, Padmanabhan S. Artificial intelligence in healthcare—the road to precision medicine. J Hosp Manag Health Policy 2021;5:29.

